Sequencing
One of the major methods of DNA sequencing in known as chain termination sequencing, dideoxy sequencing, or Sanger sequencing after its inventor, biochemist Frederick Sanger.
 The method is elegantly simple.
The method is elegantly simple.
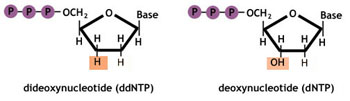
While DNA chains are normally made up of deoxynucleotides (dNTPs), the Sanger method uses dideoxynucleotides.
Dideoxynucleotides (ddNTPs) are missing a hydroxy (OH) group at the 3' position. This position is normally where one nucleotide attaches to another to form a chain. If there is no OH group in the 3' position, the additional nucleotides cannot be added to the chain, thus interrupting chain elongation.
Manual sequencing
1. Begin the process by synthesizing a chain that is complimentary to the template you want to analyze.
2. Add a specific primer that you know will anneal.
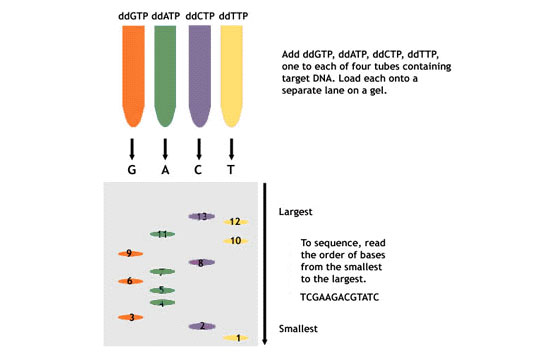
3. Divide your sample among four test tubes. One test tube will be used for each specific nucleotide (dGTP, dATP, dCTP, and dTTP).
4. Add DNA polymerase to each tube and one specific nucleotide per tube.
5. Add ddNTPs to all four tubes. Once you add the ddNTPs, there is no way for the chain to keep elongating, hence you have dd chain termination.
6. Run this on a polyacrylamide gel using one lane per reaction tube (dGTP, dATP, dCTP, and dTTP).
7. To sequence, read the order of bases from the smallest to the largest.
Automated sequencing
If this looks like too much work, you can pay for automated sequencing where a machine does most of the work for you. Of course, you'll need about $100,000 for the machine.
An automated sequencer runs on the same principle as the Sanger method (dideoxynucleotide chain termination). A laser constantly scans the bottom of the gel, detecting bands as they move down the gel. Where the manual method uses radioactive labeling, automated sequencing uses fluorescent tags on the ddNTPs (a different dye for each nucleotide). This makes it possible for all four reactions (dGTP, dATP, dCTP, and dTTP) to be run in one lane, so you can have huge numbers of reactions on one gel. This is a very efficient method.
It is important to remember, however, that a computer can make mistakes. Don't trust the computer. Always check your printout for accuracy. You are looking for a good signal, at least in the 100s, and proper spacing, ideally about 12. Look also for big gaps between the bases since the computer can miss bases. It may often miss a G after an A, especially after an AA.
View Citation
Bibliographic details:
- Article: Sequencing
- Author(s): Guruatma "Ji" Khalsa
- Publisher: Arizona State University School of Life Sciences Ask A Biologist
- Site name: ASU - Ask A Biologist
- Date published: April 12, 2010
- Date accessed: November 18, 2024
- Link: https://askabiologist.asu.edu/sequencing
APA Style
Guruatma "Ji" Khalsa. (2010, April 12). Sequencing. ASU - Ask A Biologist. Retrieved November 18, 2024 from https://askabiologist.asu.edu/sequencing
Chicago Manual of Style
Guruatma "Ji" Khalsa. "Sequencing". ASU - Ask A Biologist. 12 April, 2010. https://askabiologist.asu.edu/sequencing
Guruatma "Ji" Khalsa. "Sequencing". ASU - Ask A Biologist. 12 Apr 2010. ASU - Ask A Biologist, Web. 18 Nov 2024. https://askabiologist.asu.edu/sequencing
MLA 2017 Style
Be Part of
Ask A Biologist
By volunteering, or simply sending us feedback on the site. Scientists, teachers, writers, illustrators, and translators are all important to the program. If you are interested in helping with the website we have a Volunteers page to get the process started.








