Southern Blotting
Southern blotting was named after Edward M. Southern who developed this procedure at Edinburgh University in the 1970s. To oversimplify, DNA molecules are transferred from an agarose gel onto a membrane. Southern blotting is designed to locate a particular sequence of DNA within a complex mixture. For example, Southern Blotting could be used to locate a particular gene within an entire genome.
The amount of DNA needed for this technique is dependent on the size and specific activity of the probe. Short probes tend to be more specific. Under optimal conditions, you can expect to detect 0.1 pg of the DNA for which you are probing.
This diagram shows the basic steps involved in a Southern blot.
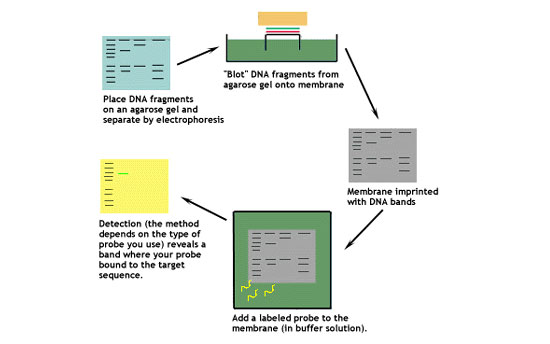
Let's look at this technique in greater detail.
1. Digest the DNA with an appropriate restriction enzyme.
2. Run the digest on an agarose gel.
3. Denature the DNA (usually while it is still on the gel).
For example, soak it in about 0.5M NaOH, which would separate double-stranded DNA into single-stranded DNA. Only ssDNA can transfer.
A depurination step is optional. Fragments greater than 15 kb are hard to transfer to the blotting membrane. Depurination with HCl (about 0.2M HCl for 15 minutes) takes the purines out, cutting the DNA into smaller fragments. Be aware, however, that the procedure may also be hampered by fragments that are too small.
Be sure to neutralize the acid after this step, or the base after the prior step if you don't depurinate.
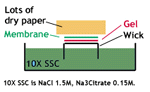 4. Transfer the denatured DNA to the membrane. Traditionally, a nitrocellulose membrane is used, although nylon or a positively charged nylon membrane may be used. Nitrocellulose typically has a binding capacity of about 100µg/cm, while nylon has a binding capacity of about 500 µg/cm. Many scientists feel nylon is better since it binds more and is less fragile. Transfer is usually done by capillary action, which takes several hours. Capillary action transfer draws the buffer up by capillary action through the gel an into the membrane, which will bind ssDNA.
4. Transfer the denatured DNA to the membrane. Traditionally, a nitrocellulose membrane is used, although nylon or a positively charged nylon membrane may be used. Nitrocellulose typically has a binding capacity of about 100µg/cm, while nylon has a binding capacity of about 500 µg/cm. Many scientists feel nylon is better since it binds more and is less fragile. Transfer is usually done by capillary action, which takes several hours. Capillary action transfer draws the buffer up by capillary action through the gel an into the membrane, which will bind ssDNA.
You may use a vacuum blot apparatus instead of capillary action. In this procedure, a vacuum sucks SSC through the membrane. This works similarly to capillary action, except more SSC goes through the gel and membrane, so it is faster (about an hour). (SSC provides the high salt level that you need to transfer DNA.)
After you transfer your DNA to the membrane, treat it with UV light. This cross links (via covalent bonds) the DNA to the membrane. (You can also bake nitrocellulose at about 80C for a couple of hours, but be aware that it is very combustible.)
5. Probe the membrane with labeled ssDNA. This is also known as hybridization.
Whatever you call it, this process relies on the ssDNA hybridizing (annealing) to the DNA on the membrane due to the binding of complementary strands.
Probing is often done with 32P labeled ATP, biotin/streptavidin or a bioluminescent probe.
A prehybridization step is required before hybridization to block non-specific sites, since you don't want your single-stranded probe binding just anywhere on the membrane.
To hybridize, use the same buffer as for prehybridization, but add your specific probe.
6. Visualize your radioactively labeled target sequence. If you used a radiolabeled 32P probe, then you would visualize by autoradiograph. Biotin/streptavidin detection is done by colorimetric methods, and bioluminescent visualization uses luminesence.
32P labeled ATP
Treat the dsDNA fragment that you are using as a probe with a limiting amount of Dnase, which causes double-stranded nicks in DNA. Add 32P, dATP, and other dNTPs to DNA polymerase I, which has 5' to 3' polymerase activity and 5' to 3' exonuclease activity.
 Nick translation occurs and as the nick is translated down the DNA strand, the polymerase activity continues to nick while the exonuclease activity continues to fill in the nick. As this happens, 32P becomes incorporated into, and thus labels, the DNA. Heat the DNA to make it single stranded, then immediately place it on ice to keep the two strands from reannealing to each other. (If the DNA is on ice, the DNA passes through the annealing temperature too quickly for the DNA to rehybridize into double-stranded DNA.)
Nick translation occurs and as the nick is translated down the DNA strand, the polymerase activity continues to nick while the exonuclease activity continues to fill in the nick. As this happens, 32P becomes incorporated into, and thus labels, the DNA. Heat the DNA to make it single stranded, then immediately place it on ice to keep the two strands from reannealing to each other. (If the DNA is on ice, the DNA passes through the annealing temperature too quickly for the DNA to rehybridize into double-stranded DNA.)
Prehybridization
To prehybridize, add non-specific ssDNA. Somicated salmon sperm DNA is commonly used. Add 20X SSC, Denhardt's solution (ficol and PVP, which are large molecules to take up space and generate more contact; and BSA, bovine serum albumen, a non-specific protein), SDS (sodium dodecyl sulfate), and formamide.
Altering the concentrations of formamide, SSC, and SDS affects "stringency," or specificity. If you have a higher stringency you should also have a higher degree of similarity between the probe and the target sequence.
View Citation
Bibliographic details:
- Article: Southern Blotting
- Author(s): Guruatma "Ji" Khalsa
- Publisher: Arizona State University School of Life Sciences Ask A Biologist
- Site name: ASU - Ask A Biologist
- Date published: April 12, 2010
- Date accessed: November 17, 2024
- Link: https://askabiologist.asu.edu/southern-blotting
APA Style
Guruatma "Ji" Khalsa. (2010, April 12). Southern Blotting. ASU - Ask A Biologist. Retrieved November 17, 2024 from https://askabiologist.asu.edu/southern-blotting
Chicago Manual of Style
Guruatma "Ji" Khalsa. "Southern Blotting". ASU - Ask A Biologist. 12 April, 2010. https://askabiologist.asu.edu/southern-blotting
Guruatma "Ji" Khalsa. "Southern Blotting". ASU - Ask A Biologist. 12 Apr 2010. ASU - Ask A Biologist, Web. 17 Nov 2024. https://askabiologist.asu.edu/southern-blotting
MLA 2017 Style
Be Part of
Ask A Biologist
By volunteering, or simply sending us feedback on the site. Scientists, teachers, writers, illustrators, and translators are all important to the program. If you are interested in helping with the website we have a Volunteers page to get the process started.








